I’ve been looking for a simple and reliable access to public health data for a while now. Eventually, I bumped into the WHO package, which allows downloading directly from the World Health Organization’s Global Health Observatory in a dynamic and reproducible way. The data is accessible after you installed the package either from the CRAN or GitHub.
# From CRAN
install.packages("WHO")
# From Github
library(devtools)
install_github("expersso/WHO")Nothing fancy and the rest is pretty straightforward too: only two functions necessary.
- the
get_codesto return a dataframe with series, codes, and descriptions for all available series - the
get_datato retrieve the data from the internet and create directly your dataframe
How to retrieve series from the WHO
# Retrieve series, codes, and descriptions
codes <- get_codes()
str(codes)
## tibble [3,316 × 3] (S3: tbl_df/tbl/data.frame)
## $ label : chr [1:3316] "MDG_0000000001" "MDG_0000000003" "MDG_0000000005" "MDG_0000000006" ...
## $ display: chr [1:3316] "Infant mortality rate (probability of dying between birth and age 1 per 1000 live births)" "Adolescent birth rate (per 1000 women aged 15-19 years)" "Contraceptive prevalence (%)" "Unmet need for family planning (%)" ...
## $ url : chr [1:3316] "https://www.who.int/data/gho/indicator-metadata-registry/imr-details/1" "https://www.who.int/data/gho/indicator-metadata-registry/imr-details/4669" "https://www.who.int/data/gho/indicator-metadata-registry/imr-details/5" "https://www.who.int/data/gho/indicator-metadata-registry/imr-details/6" ...So far they are 3316 datasets available, which are all easily retrieved through the get_data function. But first we need to pick-up series of interest. Let’s say we want to analyze breast cancer data and search for them among the series with a regular expression.
# Search for series about breast cancer
breastCancer <- codes[grep("[Bb]reast [Cc]ancer", codes$display),]
breastCancer$display
## [1] "Age-standardized DALYs, breast cancer, per 100,000"
## [2] "Age-standardized death rates, breast cancer, per 100,000"
## [3] "General availability of breast cancer screening (by palpation or mammogram) at the primary health care level"
## [4] "Existence of national screening program for breast cancer"
## [5] "Age-standardized DALYs, breast cancer, per 100,000"
## [6] "Age-standardized death rates, breast cancer, per 100,000"So we have age-standardized disability-adjusted life years (DALYs), age-standardized death rates, and general availability of breast cancer screening at the primary health care level there. Certainly enough to run some test analyses. Okay let’s first fetch the data through the get_data function.
# Retrieve the dataframes from the internet
DALYs <- get_data(breastCancer$label[1])
deathRates <- get_data(breastCancer$label[2])
cancerScreening <- get_data(breastCancer$label[3])Interlude
Maybe you are relatively new to R. If you recently installed it on your computer and didn’t have time to explore the CRAN you might want to run the following code to ensure you have all the required packages installed. All of them are very useful anyway: you won’t regret it!
# Required packages from CRAN
.pkgs = c("dplyr", "ggplot2", "RColorBrewer")
# Install required packages from CRAN (if not)
.inst <- .pkgs %in% installed.packages()
if(length(.pkgs[!.inst]) > 0) install.packages(.pkgs[!.inst])After that, be sure to load them all.
# Load required packages
library(dplyr)
library(ggplot2)
library(RColorBrewer)Let’s create our dataframe
After loading the data, we surely want to combine our three dataframes together. Male breast cancers are relatively rare, about 1% of all breast cancers only and are usually diagnosed at a more advanced stage. Therefore, I choose to filter them out and to return here the female breast cancers only.
# Filter, combine and group together
df <- data.frame(deathRates %>%
filter(sex == "Female") %>%
group_by(year, country) %>%
summarise(region, value),
DALYs %>%
filter(sex == "Female") %>%
group_by(year, country) %>%
summarise(value),
cancerScreening %>%
filter(country %in% DALYs$country) %>%
group_by(year, country) %>%
summarise(value))There is some redundancy in the country and year columns that needs to be removed. A simple way to do that is to use a regular expression again. Once you’ve selected the redundant columns, it becomes easy to clean the dataframe.
# Search and remove redondancy
sel.cl <- grep("*[yr].[12]", colnames(df), invert = TRUE)
df <- df[,sel.cl]Finally, let’s quickly adjust the column’s names of our dataframe.
# Rename columns
colnames(df) <- c("year", "country", "region", "deathRates", "DALYs", "cancerScreening")
df[1:10,]
## year country region deathRates DALYs
## 1 2004 Afghanistan Eastern Mediterranean 29.6 506
## 2 2004 Albania Europe 28.7 384
## 3 2004 Algeria Africa 17.5 212
## 4 2004 Andorra Europe 18.4 267
## 5 2004 Angola Africa 34.5 410
## 6 2004 Antigua and Barbuda Americas 37.7 549
## 7 2004 Argentina Americas 25.8 316
## 8 2004 Armenia Europe 38.6 552
## 9 2004 Australia Western Pacific 20.3 337
## 10 2004 Austria Europe 20.1 259
## cancerScreening
## 1 Yes
## 2 Yes
## 3 Yes
## 4 Yes
## 5 No data received
## 6 Yes
## 7 Yes
## 8 Yes
## 9 Yes
## 10 YesNow what?
Well, how about plotting the data now?
ggplot(df, aes(x = deathRates, y = DALYs, color = region, shape = cancerScreening)) +
geom_point() +
theme_minimal() +
ggtitle("") +
xlab("Death rates (per 100,000)") +
ylab("DALYs (per 100,000)") +
scale_shape_manual(values = c(1:5), name = "Screening") +
scale_color_brewer(palette = "Set1", name = "Region") +
theme(legend.position = "bottom") +
theme(legend.title = element_blank())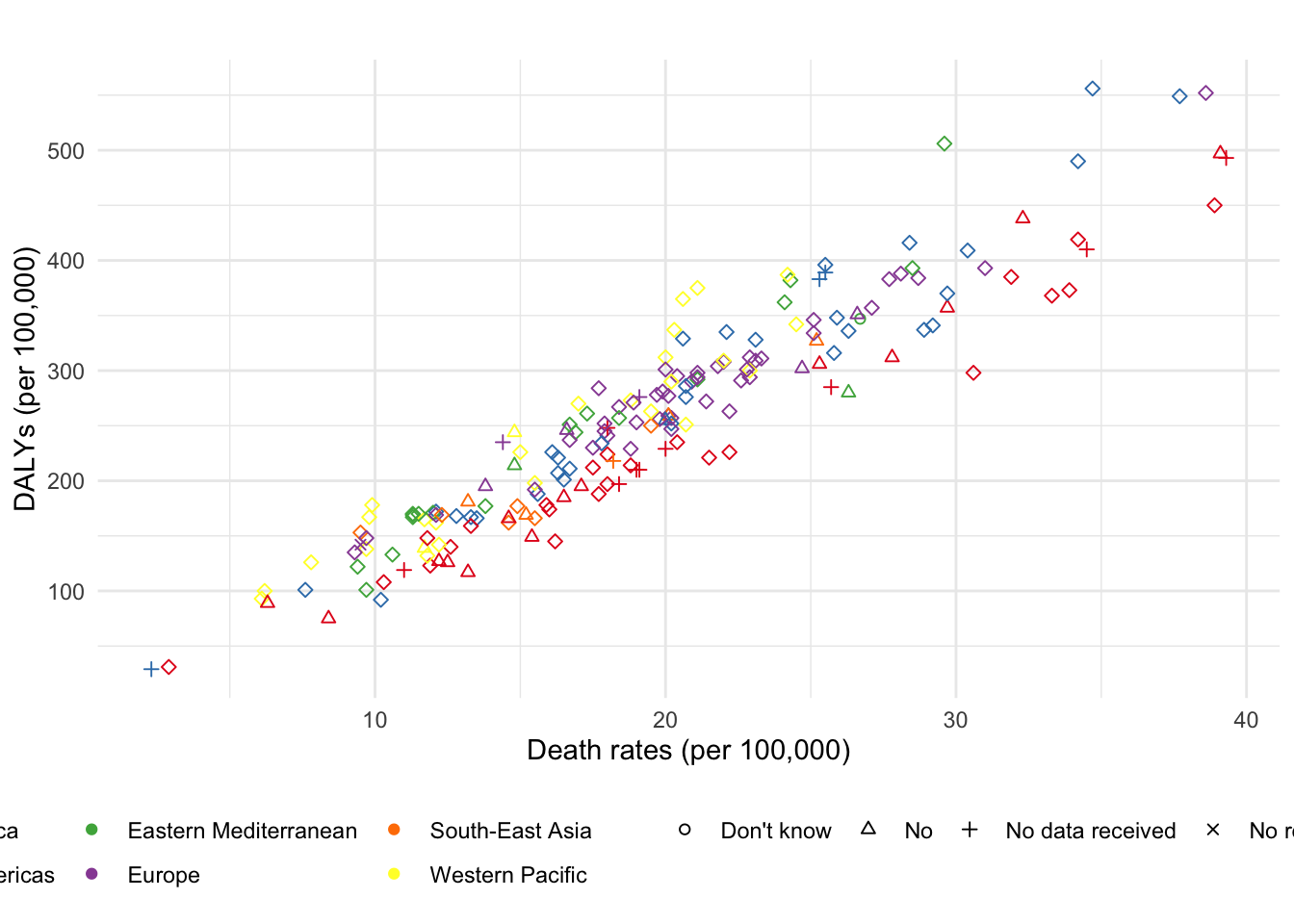
Figure 1: A fancy scatterplot.
Two assumptions were made in figure 1: death rates and DALYs were published in 2004, whereas the data about the availability of screening is from 2013. We can reasonably assume that countries with no screening at the primary health care level in 2013 didn’t have screening back in 2004 either. But there is no way to be sure that countries organizing screening for breast cancer nowadays were already doing it in 2004. Additionally, there is no data about patient’s sex in the breast cancer screening data set. But one can assume that it was a least available to female patients, since they are usually the only population targeted by routine screening for breast cancer.
